
|
|
Main /
TutorialsThese tutorials are out of date, please see http://cmic.cs.ucl.ac.uk/camino for current tutorials and documentation.While most of the current code is backwards compatible, some of the commands here may no longer work. Please see the current tutorials at the new site before emailing us with problems. Thanks.Input data to Camino Conversion of raw data from DICOM to NIfTI NIfTI conversion to Camino raw format Dealing with diffusion tensors in NIfTI format Detecting Crossing Fibres in the post-mortem pig brain 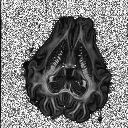 Creating FA map Creating FA map 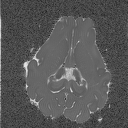 Creating Tr(D) map Creating Tr(D) map 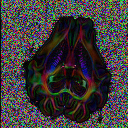 Creating colour-coded direction map Creating colour-coded direction map  Detecting fibre crossings Detecting fibre crossings
Create a Q-Ball ODF map (grey scale and color) Extracting useful information about the peaks of the Q-Ball ODFs Visualising Q-Ball ODF fibre-orientation estimates
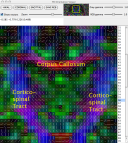 Viewing the colour-coded direction map with pdview Viewing the colour-coded direction map with pdview  Using an ROI Using an ROI 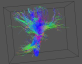 Streamline Tractography Streamline Tractography  Probabilistic Tractography using PICo. Probabilistic Tractography using PICo. 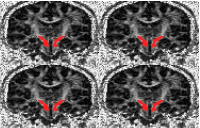 Parallel processing of PICo probability maps. Parallel processing of PICo probability maps.Tractography with q-ball and PASMRI Running reduced-encoding PASMRI How to construct a connectivity matrix  PICo using waypoints and target ROIs PICo using waypoints and target ROIs 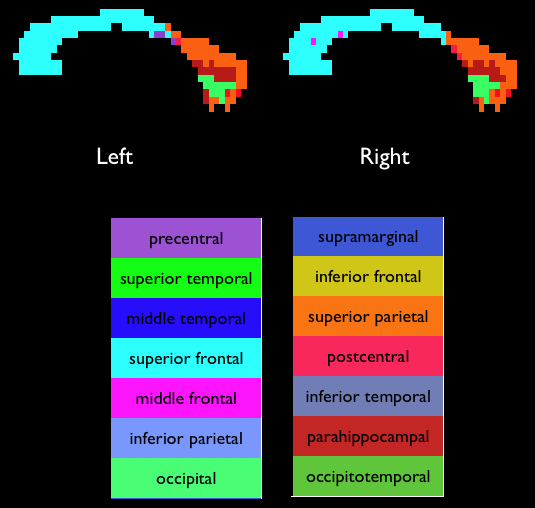 Segmenting brain structures by connectivity Segmenting brain structures by connectivity Running PASMRI using parallel processing Running PASMRI using SGE Creating PAS plots. Using camino to produce synthetic data Synthetic data from 3D Gaussian functions Synthetic data using Monte-Carlo simulation Generating statistics on fibre orientation estimates Constructing data files using shredder Synthetic data for fibre-crossing studies Spatial normalization and warping of tensor images Creating masked diffusion tensor images Using flirt to compute affine transformations between images Warping a diffusion tensor image Reorienting diffusion tensors for consistency with the image transformation
Creating a general-waveform schemefile Mapping axon density and diameter with ActiveAx |