Evolutionary Drug Discovery
EPSRC project GR/S03546/01
with
GSK
on using genetic programming in drug discovery.
This follows the
INTErSECT
project where we automatically evolved
combined classifiers
by fusing together many diverse classifiers.
We have used genetic programming to predict interaction between
chemicals (proto-drugs)
and human cell wall enzymes.
Training data being provided by both
high throughput screening (HTS)
and IC50 measurements at GSK.
Recent work include modelling bioavailability and gene expression
networks.
Project Presentations
-
Cercia workshop on Computational Intelligence in Cheminformatics,
2 March 2006
-
British Computer Society
12 Feb 2004
Slides.
Handout.
-
Invited talk,
European Workshop on Data Mining and Text Mining for Bioinformatics,
Dubrovnik, Croatia,
22 September, 2003.
- Seminar,
Essex University, 20 August 2003.
- BioGEC'2003
workshop, Chicago, 12 July 2003.
- Transferring Computer Science Research to
Mining DNA chip Protein Expression.
Talk and
poster presentation at
MIPNETS
25-27 June 2003.
- Seminar
Colorado State University, 19 May 2003.
- Invited talk
PRCVC Prague, April 2003.
(Abstract).
- Invited talk
Hybrid Information Systems, Santiago, 2002.
- Invited talk
IBERAMIA, Seville, 2002.
Slides.
-
Knowledge Discovery meets Drug Discovery,
Leuven, 23 Oct 2002.
Project Research Papers
Code
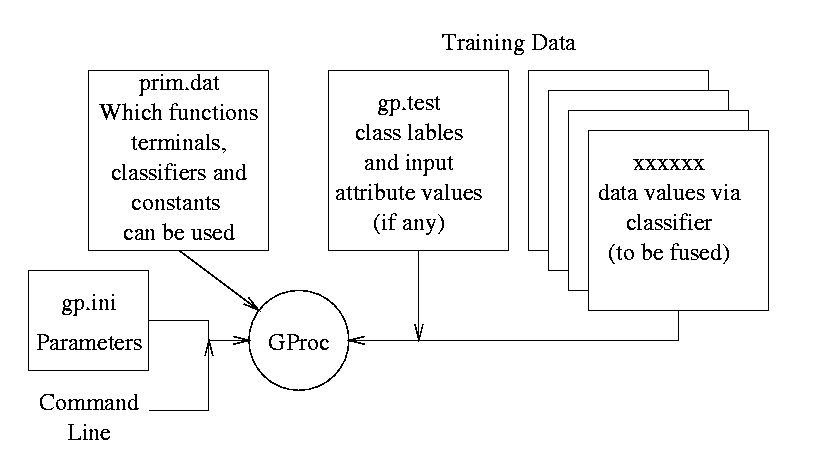
The genetic programming system GProc
is controlled by parameters given by a file gp.ini.
However these can be overridden via the command line.
The terminal and function sets are given by the contents of file
prim.dat
Training data is given in gp.test.
When working in classifier fusion mode,
one or more additional files (which are specified by prim.dat)
contain the output predictions (and the classifier confidence in it).
Output is directed to standard output.
Version 1.8b
W.B.Langdon
6 June 2003
(last update 4 Oct 2012)

