Tutorial: Motion Correction for Diffusion Weighted Images
This tutorial introduces the function mbalign in Camino, which is used to align the diffusion-weighted images within a single acquisition.

SYNOPSIS
mbalign [options]
1. Preparation before running mbalign
1.1.General data files and information
Before running mbalign, we need to have input and scheme files ready, and use
-inputfile <Input voxel-order file>
-schemefile <Scheme file name>
to specify in command options. If the input file is in scanner-order, we can use
-scanner -inputfile <Input scanner-order file>.
And also we need to make some other information of input image data clear and specify a few other options
-datadims X Y Z <Number of voxels in each dimension>
-voxeldims x y z <Voxel size in mm>
-sigma <Standard deviation of noise>
1.2.Sigma
The value of sigma is the approximate noise standard deviation. An estimate of sigma is sqrt(E(S^2)/2), where S is the signal in background and E denotes expectation over an ROI. The camino program, datastats, can work it out for you:
datastats -schemefile S.scheme -bgmask S32_BG.Bshort -inputfile S32.Bfloat
where S32_BG.Bshort is a binary image containing an ROI in a background region of the image.
The output looks like this:
=============================================================================================
Foreground voxel count: 142584
Component E(S) E(S^2) Var(S) Std(S)
1 2.882503E02 9.834995E04 1.526169E04 1.235382E02
2 3.010773E02 1.070645E05 1.641700E04 1.281288E02
3 2.876059E02 9.854322E04 1.582610E04 1.258018E02
4 2.564062E02 9.868863E04 1.294450E04 1.137739E02
5 2.017598E02 9.636593E04 1.028376E04 1.293741E02
6 2.915845E02 9.591012E04 1.817574E04 1.197654E02
:
:
:
==============================================================================================
A well chosen background region that genuinely contains no signal should show similar statistics in the output above in both non-diffusion-weighted (b=0) images and diffusion weighted images. The first four images in the example above have b=0 and the subsequent ones have b>0. We see little difference in the statistics, which suggests the background genuinely contains no signal.
The value of E(S^2) is around 1E5, so we might pick sigma = sqrt(E(S^2)/2), ie around 224.
However, you may need to play with the setting of sigma to get the best results out of mbalign. The value is somewhat artificial, because really we just seek a good
threshold that reliably rejects measurements from corrupted images from contributing to the diffusion tensor fit. Several users have reported that they have to set
sigma 100 or 1000 times smaller than the estimate of sigma described above to get good results. Others report having to set sigma much higher, so this seems to be
data dependent.
1.3.Make slice to check input volume
This section shows a simple way to check the input volume is what the program expects and also to give you a fell for how much realignment is required. It creates an image volume containing the corresponding slice from each image volume in the data set.
If the image file is in voxel-order, we need to transfer it to scanner-order first:
voxel2scanner -voxels $((128*128*32)) -inputdatatype float -outputdatatype float -components 64 -inputfile S32.Bfloat > S32.scan.Bfloat
Then we can use camino function shredder to extract one slice from each of the 64 3D components, and build a 3D image shown in Fig1:
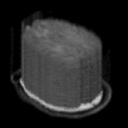
Fig.1 An image volume containing 1 slices from each of the 64 component images in the data set.
Here is an example script for creating the image volume above:
COMPONENT=64
DATADIM_X=128
DATADIM_Y=128
DATADIM_Z=32
#Since our dataset used in the example is float, so
TYPESIZE=4
# Extract the middle slice
OFFSET=$(($DATADIM_X*$DATADIM_Y*$((DATADIM_Z/2))*$TYPESIZE))
shredder $OFFSET $(($DATADIM_X*$DATADIM_Y*$TYPESIZE)) $(($DATADIM_X*$DATADIM_Y*$(($DATADIM_Z-1))*$TYPESIZE)) < S32.scan.Bfloat > S32._SLICECHECK.img
# Make a header file
VOXELDIM_X=1.88
VOXELDIM_Y=1.88
VOXELDIM_Z=2.0
analyzeheader -voxeldims $VOXELDIM_X $VOXELDIM_Y $VOXELDIM_Z -datadims $DATADIM_X $DATADIM_Y $COMPONENT -datatype float > S32._SLICECHECK.hdr
Now we can use visualisation tools, like MRIcro (Fig.2), to check the alignment of input data set.
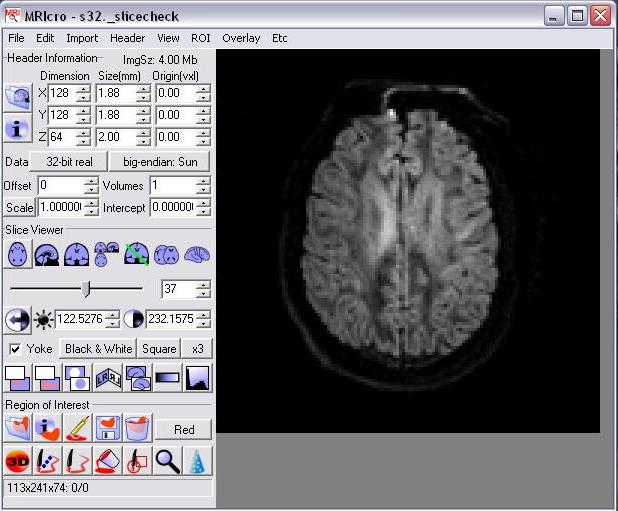
Fig.2 Using MRIcro to see slice volume
2. Run mbalign
2.1. Run mbalign simply
An example of using mbalign in the simplest way is:
mbalign -datatype float -schemefile S.scheme -datadims 128 128 60 -voxeldims 1.88 1.88 2.0 -sigma 1.9 -inputfile S32.Bfloat
then the screen output would be:
=============================================================================================
:-) I have everything I need!
WARNING: No -bgmask and -bgthresh input, using zero background threshold. Performance may improve with better threshold.
WARNING: No -components input. Using 64 components from schemefile. If wrong, restart with -components option.
64 components inside inputfile.
No temp directory specified. Trying to use /tmp/S32_17.03.08_180229.
Successfully created /tmp/S32_17.03.08_180229.
Linux system detected.
No output file name specified. Output file will be: /tmp/S32.out.Bfloat
Disk space temporarily used during calculation is about 2264924160. Make sure space is available!
==============================================================================================
The program will create a temporary directory used for calculation process. It can be either specified by -tmpdir or automatically create according to the file name and current time.
We do not have to use -outputfile to specify the output file name, and the program can generate it according to the input file name, which is like the example shown above.
To run mbalign, computer need to have registration software flirt(http://www.fmrib.ox.ac.uk/fsl/flirt/index.html) installed, which is part of FSL library(http://www.fmrib.ox.ac.uk/fsl/). We also need to specify the flirt direction by using -fsldir, but more easily, once we have camino and FSL installed, default value of variable DIR_FSL can be set which can be easily found inside mbalign.
During the running of mbalign, there could be a lot of warning messages showing inside terminal window. Normally, just do not worry about it too much. Once the registration is complete, we may see the message shown below.
==============================================================================================
...
Registering ori.ScannerOrder.Bfloat.ck by using ref.ScannerOrder.Bfloat.ck
Registering ori.ScannerOrder.Bfloat.cl by using ref.ScannerOrder.Bfloat.cl
Transferring output to Big-endian format...
Making img and hdr files for slice checking...
Transfer output file to voxel-order...
Removing junk files...
Scheme file not updated.
Aligned data set output to /tmp/S32.out.Bfloat.
Program finished at
Mon Mar 24 23:36:24 GMT 2008
==============================================================================================
After program finishes, the temporary directory would be deleted (Removing junk files...), but we can still use -keepjunk to make it kept, which could be useful to help us to analyse the final output. Certainly, if the program is interrupted, this temporary fold will remain as junk files in computer.
If we did not specify -outputfile, the program can generate it according to the input file name (Aligned data set output to /tmp/S32.out.Bfloat).
We will discuss scheme file updating in Section 4.Update Gradient.
2.2. Run mbalign in an advanced way
Some other options mignt need to use:
-flirtsearchcost <Search cost function used in flirt>
Default cost function is mutualinfo (Mutual Information). Other options are corratio,normcorr,normmi,leastsq.
-flirttransform <Transformation used in flirt>
Default transformation is affine. The other option is rigid.
-searchrange <angle>
Default is 90, which means search range is between -90 and 90 in all x, y and z directions.
-eddy
Specifies registration for eddy-current induced distortion.
-datatype <Data type for input and output files>
Default is float.
-scanout <output scanner-order file>
Adds an extra output file in scanner-order. This won't stop default voxel-order output.
-omat <File name>
Output transform matrix in ascii format.
-slicecheck <File name>
Output a pair of <File name>.img and <File name>.hrd files. Default is no calculation.
When all the options are decided, we can run mbalign in an advanced way, such like
mbalign -datatype float -schemefile S.scheme -datadims 128 128 60 -voxeldims 1.88 1.88 2.0 -sigma 1.9 -fsldir /cs/research/medim/common0/green/common/fsl/fslRH9/ -inputfile S32.Bfloat -slicecheck S32.fmr.slice.check -outputfile S32.fmr.Bfloat -omat S32.fmr.mat.txt -scanout S32.fmr.scanout.Bfloat -keepjunk -tmpdir tmp.fmr
3. Improve performance
There are a few options can be used to improve the performance of mbalign.
-sigma
High sigma allows the program to involve more measurement in the DT fitting, and low sigma leads rejections during the DT fitting. Based on this theory, we can change the value of sigma to improve the reference making.
-bgmask <Mask file>
Use a mask file can improve the quality of the reference images used in mbalign registration. And the data type of mask file should be "short".
Camino function mask can help to create a background mask from a voxel-ordered DW data file by thresholding the average b=0 measurement.
mask -inputfile S32.Bfloat -inputdatatype float -schemefile S.scheme -bgthresh 100 -outputdatatype short > S32_M100.Bshort
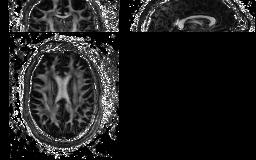
Fig.3 View projection of FA map generated by camino

Fig.4 View projection of mask file generated by camino
We can also use matlab to make a mask file.
-bgthresh <Background threshold>
Decide the value of threshold, and improve fitting the diffusion tensors.
-searchrange <angle>
Sometimes, the pitch of histogram will cause the failure of registration. Simply narrow down the angle search range can cover this problem for most of the time.
4. Update Gradient
Updating diffusion gradients after registration is not an essential procedure, but can improve the registration result. Fig.5 explains the reason why diffusion gradients need to be updated after registration.
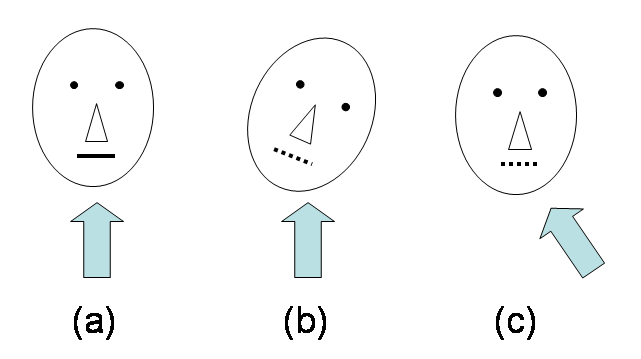
Fig.5 Illustration of how rotation affects the effective gradient direction. The arrows indicate gradient directions. (a) Head without rotation. Suppose the mouth is a fibre. The signal is high because the gradient is perpendicular to the fibre. (b) Head with rotation. The signal is lower in the mouth fibre. (c) The unrotated head (after registration). The effective gradient direction for the corrected image is rotated. We can use our matlab function to update diffusion gradient for registered data set, and the usage method is explained inside the matlab file. Since the transformations used in the registration contribute to the gradient updating, we MUST save the transform matrix when running mbalign, using -omat.
5. More hints
Quite a few default options can be change in the source code of mbalign. Make the default values to the ones most frequently used can make the everyday use of mbalign much simpler. Inside mbalign source code, we can find and change the default options from the following part.
####################################################
##### Change default variables to match system #####
####################################################
# Hint: To make your input arguments simple, set
# default input which you most often to use.
# FSL directory
DIR_FSL=/cs/research/medim/common0/green/common/fsl/fslRH9
# LIM_ROTATE is default for -searchrange
LIM_ROTATE=90
# Available cost functions are:
# mutualinfo corratio,normcorr,normmi,leastsq.
SEARCH_COST=mutualinfo
#Degree of freedom
# 12 for affine; 6 for rigid.
DOF=12
Acknowledgement
We would like to thank Geoff Parker and Karl Embleton, University of Manchester, for providing the brain data.
Reference
- Y. Bai and D. C. Alexander, “Model-Based Registration to Correct for Motion between Acquisitions in Diffusion MR Imaging”, The Fifth IEEE International Symposium on Biomedical Imaging ( ISBI 2008), May 2008.